Fungi have always been an important source of biologically active natural products. The modern age of antibiotics was ushered in by the discovery of penicillin, isolated from molds of Penicillium. Since then, many blockbuster drugs have been fully or partially derived from fungal sources including the cephalosporins, statins, and ciclosporins, to name just a few. It is no surprise then that researchers are still unearthing new natural products from fungi. What is surprising is that very few RiPPs (ribosomally synthesized and post-translationally modified peptide natural products) have been found in fungi. Our lab loves RiPPs! RiPPs are commonly produced by bacteria, especially the “soil-lovers” like Bacillus and Streptomycetes, in which lantipeptides, thiopeptides, sactipeptides, thiazole/oxazole modified microcins (TOMMs) and other RiPP biosynthetic gene clusters are sprinkled throughout the genomes. In fungi, on the other hand, only a handful of RiPPs are known. The larger size and complexity of fungal genomes or the often the toxic nature of their metabolites may be responsible for this gap, but either way, given the rich biology of both RiPPs and fungal natural products, fungal RiPPs should be interesting.
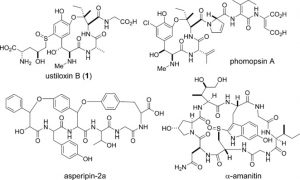
This logic heightened our interest in a fairly recent series of papers on the ustiloxins and phomopsins, which have been newly identified as RiPPs from filamentous fungi. These are not new compounds. They were discovered separately back in the 80’s and 90’s, and Tom Wandless did beautiful syntheses of both ustiloxins and phomopsins in the early ‘00s. They are related by a common core, three-amino acid ring, cyclized via an aryl ether between the meta-hydroxyl of DOPA and the β-carbon of an isoleucine residue. The ustiloxins also has a novel, Cys-derived sulfoxide moiety appended to the aromatic ring of DOPA. Crystal structures have revealed that the ether macrocycle provides important contacts for the tubulin-binding activity of phomopsins. Separate reports from the Oikawa lab (Hokkaido) and the labs of Wilfred van der Donk (Illinois) and Qi Zhang (Fudan) detail the identification of their biosynthetic gene clusters in the genomes of Ustilaginoidea virens (ustiloxin) and Phomopsis stromiformis (phomopsin), respectively. The two biosynthetic clusters bear remarkable similarities, including the sequence of the core peptide: both ustiloxins and phomopsins are biosynthesized from the same primary sequence ‘V-E-D-Y-x-I-x-x-D/N-K-R’. While the phomopsins retain all residues of the Y-x-I-x-x-D/N motif in the final structure, the ustiloxins retain only the Y-x-I-x residues. This may indicate a potential truncation of the isolated ustiloxins or may point to certain fundamental differences in the biosynthetic enzymatic reactions, structure-activity relationships and physiological targets of the two fungal toxins. Both reports generate a series of knockouts and reconstitute some of the tailoring enzymes in order to validate the cluster identifications. Zhang and van der Donk were also able to identify 27 similar biosynthetic gene clusters in other fungi by using the sequences of the precursor peptide and the key biosynthetic enzymes. This work was leveraged to demonstrate the production of phomopsin-like molecules from several of these strains, suggesting that fungal RiPPs may be still more widely distributed than first thought.
These findings lead to several exciting questions. First and foremost, what is the enzymatic basis of the aryl ether formation? Both reports implicate the tyrosinase or ‘Q-enzyme’, but this would be a unique activity for a class of enzymes that normally catalyzes oxidation of tyrosine to DOPA. Phyre models don’t begin to indicate any additional domain on the Q-enzymes that might activate the isoleucine β-carbon. An alternative route might involve the somewhat enigmatic ‘Y-enzymes,’ which are shared by both pathways, but bear little or no homology to any known enzymes. Indeed, these Y-enzymes might hold the key to another question, that of the enzymatic basis for the novel dehydroamino acids at the C-terminus of the phomopsins, dehydro-aspartate and dehydro-isoleucine, as well as the olefins incorporated on several other residues. It is possible that the Y-enzymes constitute a novel family of oxidases that introduce these dehydrogenations, a kind of fungal alternative to the bacterial Lan-dehydratases, capable of modifying a broader variety of residues. Perhaps a similar dehydro-amino acid is installed at the isoleucine that forms the macrocycle juncture, serving as an electrophile for the ether forming reaction. This potentially novel RiPP enzymology points to the untapped potential of fungal RiPPs and the possibilities for discovering new molecules and new enzymatic reactions. It also underscores the need for further exploration of fungal genomes and the development of fungi-focused bioinformatics tools. Further studies will undoubtedly yield interesting insights and potentially novel therapeutics.
