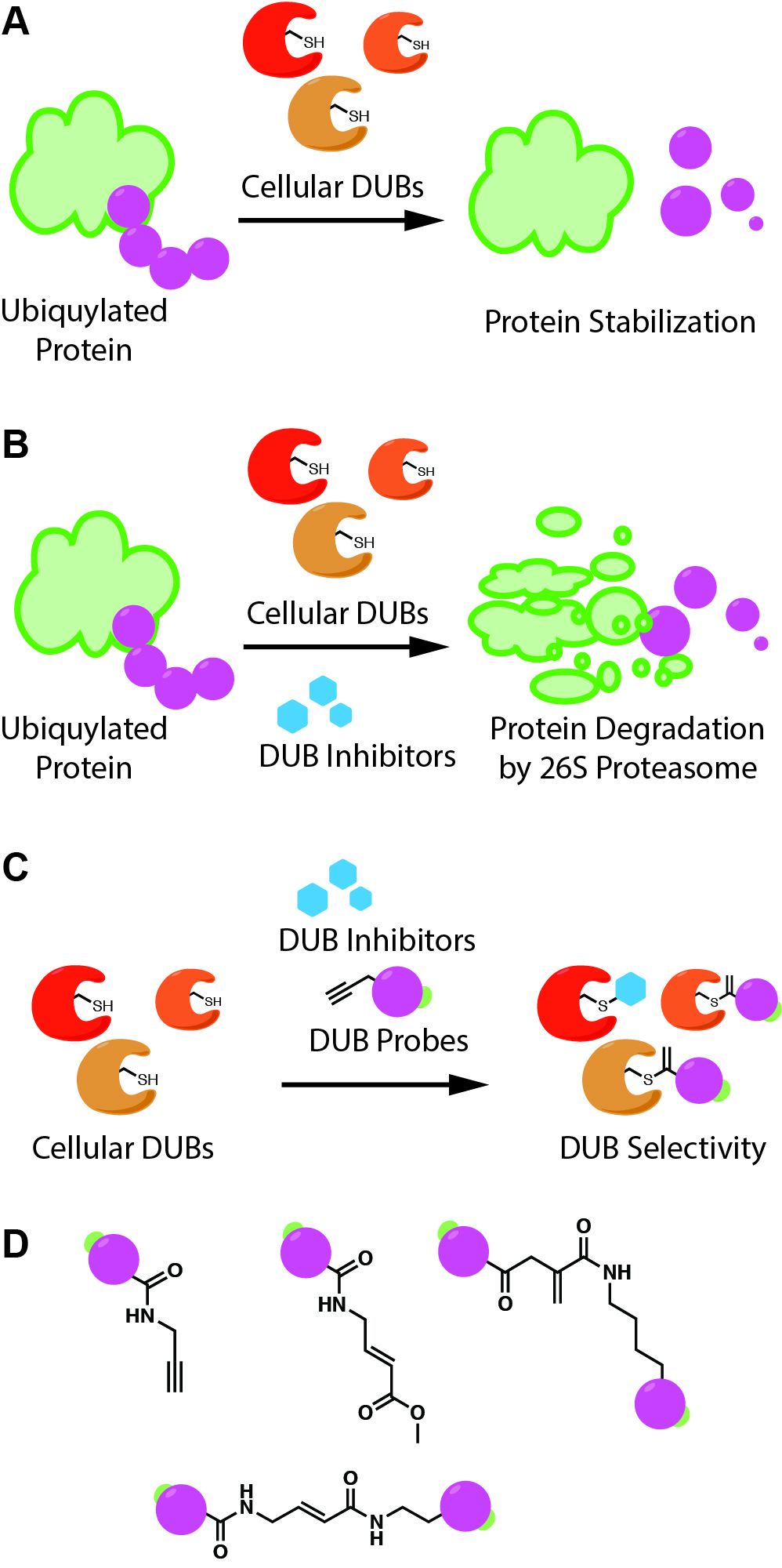
Ubiquitin (Ub) is ubiquitous. This small, 76-residue protein is used to mark cellular proteins for degradation by the ubiquitin proteasome system (UPS). Two key enzyme families, Ub ligases and deubiquitylation enzymes (DUBs), are responsible for putting on and taking off these Ub units and controlling the overall protein population of the cell. As you might imagine, this dynamic process is important, and dysregulation can lead to an array of bad things. Thus, understanding the relations between DUB or Ub ligase activity and specific diseases would be helpful in designing inhibitors or other therapeutic interventions. Accordingly, a number of Ub-based probes for activity-based protein profiling (ABPP) have been developed, typically involving an affinity tag, such as hemagglutinin (HA) or biotin, on the N-terminus and an electrophilic warhead on the C-terminus, such as propargylamide (PA), vinyl methyl ester (VME), or dehydroalanine (Dha). However, many of these probes are limited to experiments with purified recombinant proteins or cell lysates due to poor cell penetrance. Probes really need to be able to enter cells and react with enzymes intracellularly in order to gain the most useful information for drug targeting and other efforts.
To this end, the Zhuang group at the University of Delaware set out to discover a cell penetrating activity-based probe to better understand the function and regulation of DUBs inside cells. The probes consisted of a cell penetrating peptide (CPP), either a cyclic polyarginine (cR10) or TAT, and an electrophilic warhead, either PA or VME. These probes were validated using purified recombinant DUBs as well as cell lysates. Additionally, they confirmed that the probes enter cells by using a fluorescent tag and fluorescence microscopy. The authors then used the probes in HeLa cells as a general probe to see which DUBs they could tag, and as a way to evaluate the efficacy of a pan-DUB inhibitor. What they found was that the cR10 probes were more efficient at getting into cells and as a result pulled down more DUBs. However, in comparison to the TAT probe, cR10 probes pulled down slightly different DUBs than the TAT probes. Interestingly, TAT probes pulled down more DUBs that are known to be located in the nucleus of the cell, however the authors showed the cR10 pulled down DUBs found in the nuclear fraction of cell lysate. In terms of the pan-DUB inhibitor, PR-619, the authors only noticed a slight decrease in the number of DUBs the probe was able to tag in the presence of inhibitor, whereas they had expected to see a large decrease in the number of DUBs they were able to pull down. They concluded from this particular experiment that the inhibitor may not be as cell-permeable as originally thought, but that their probes could still be used to evaluate other inhibitors.
Overall, the authors convinced us that their probes are cell permeable and can be used to evaluate DUB inhibitor efficacy. Our lab discussion of the paper raised several questions. For example, we previously blogged about a paper that investigated the ability of various CPPs to enter cells and their mechanisms of action. Even so, we were still very surprised to see that CPPs were able to transport such large cargoes efficiently. We had concerns about how much of the probe is actually getting into the cytoplasm as opposed to being trapped in endosomes because many CPPs are known to enter cells through endocytosis. In terms of application, the current probes seem well-suited for assessing both potency and specificity of DUB inhibitors, but we think that they could also be used to probe linkage specificity in cells. For instance, with di-ubiquitin probes conjugated to a CPP, it would be interesting to see if results for linkage specificity in cells matches what has previously been found with purified recombinant DUBs. Finally, we thought it might be interesting to use these probes in different cell and tissue types to see how the regulation of DUBs differs. This could be useful in identifying dysregulated DUBs in certain disease states. Clearly, there is significant potential for these probes to enlighten the field by providing information and knowledge in an area where there is currently very little. Indeed, the authors predict that they will be able to use their probes to determine how DUBs function and are regulated in response to various stimuli, and to target specific organelles such as the nucleus. All in all, I don’t think we’ve seen the last of these intracellular DUB probes in chemical biology or drug discovery.
